Contents
There is no dearth of materials around the potential of CRISPR and how it is already impacting medicine, food, biomaterials—basically our everyday lives. With researchers putting their best foot forward globally to fight the recent novel coronavirus outbreak, one might question “Where is CRISPR?” Not far behind! Efforts are underway to use CRISPR-based tests for viral detection.
Particularly noteworthy in this regard are two independent efforts led by Mammoth Biosciences and CRISPR pioneer Feng Zhang. Both groups have developed easy-to-use diagnostic assays using CRISPR technology for the detection of viral RNA from samples.
Read on to know more about how CRISPR-based diagnostics are aiding the fight against coronavirus.
What is Coronavirus?
Coronavirus is a family of viruses that includes mildly infective viruses as well as those causing Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS). The one making the latest rounds, which was first reported in Wuhan province in China, is a novel coronavirus that hasn’t been previously identified. The official name of the virus is now SARS-CoV-2 and the disease is Coronavirus disease 2019 (COVID-19), according to the CDC website.
Coronavirus transmission occurs by inhaling respiratory droplets from an infected person. Moreover, viruses from the same family may largely differ in their infectivity and severity, so it is difficult to estimate the danger to public health with the new coronavirus.
As the number of cases is steadily growing across the globe with about 90,870 confirmed cases worldwide, the panic of a pandemic is growing. Amidst these concerns, researchers have been hard at work by publishing over 500 research articles on coronavirus in the past month and sequencing the genomes of viral strains to uncover the patterns underlying the spread of the disease.
Along with developing treatments, the identification of infected people is of vital and immediate importance to contain the spread of the disease. The coronavirus tests available now are based on qRT-PCR and require at least 4-6 hrs (sometimes even more) to diagnose the contagion. Because of the easy transmission of this potentially deadly disease, easy-to-use kits for quick detection of the virus in human samples are the need of the hour.
Now, CRISPR researchers have stepped up with a possible solution. Experts at Mammoth Biosciences and co-founders of SHERLOCK Biosciences have independently developed CRISPR-based platforms to detect the SARS-CoV-2 RNA from samples in 30-60 min.
How does a gene editing tool help the detection of viruses? Find out in the next section.
How is CRISPR Used in Coronavirus Detection
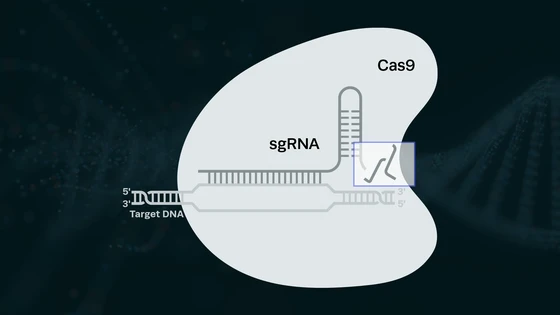
CRISPR gained popularity as a genome editing tool after its advent, but researchers soon realized its potential in diagnostics. The CRISPR mechanism relies on the guide RNA finding its complementary target sequence and the commonly used Cas 9 nuclease cutting at the precise site. Interestingly, thanks to the research on alternative Cas nucleases, CRISPR’s functionality as a location-finder, rather than site-specific cleavage tool, has emerged in the past few years.
Two nucleases, Cas12a and Cas13a, have been particularly popular in CRISPR diagnostics. While Cas12a is DNA-specific, Cas13a works with RNA. Part of the mechanism is similar to that of Cas9—a guide RNA that is complementary to the target sequence is required for specific binding and the Cas12a/Cas13a nuclease cleaves at the site. An interesting feature of Cas12 and Cas13 nucleases is that they show trans or collateral cutting activity. On finding the target, the nucleases will cut other non-targeted nucleic acid molecules in the vicinity as well. This trait is leveraged to build reporter systems for a visual readout in CRISPR diagnostics.
Using SHERLOCK Technology for Coronavirus Detection
In 2017, Feng Zhang’s group first reported a CRISPR-based nucleic acid detection technique called SHERLOCK (Specific High sensitivity Enzymatic Reporter unLOCKing). After refining it over the years, the authors published a protocol for using SHERLOCK for sensitive detection of nucleic acids in 2019. In our previous interview with Omar Abudayyeh and Jonathan Gootenberg, co-inventors of the SHERLOCK technology, the duo explained the basis of their CRISPR diagnostic platform. You can tune in to the podcast interview here.
In their latest protocol, which is freely available here, Zhang and team targeted two genes, S gene and Orf1ab, from the COVID-19 genome. In their proof-of-principle tests, the team used synthetic COVID-19 virus RNA fragments.
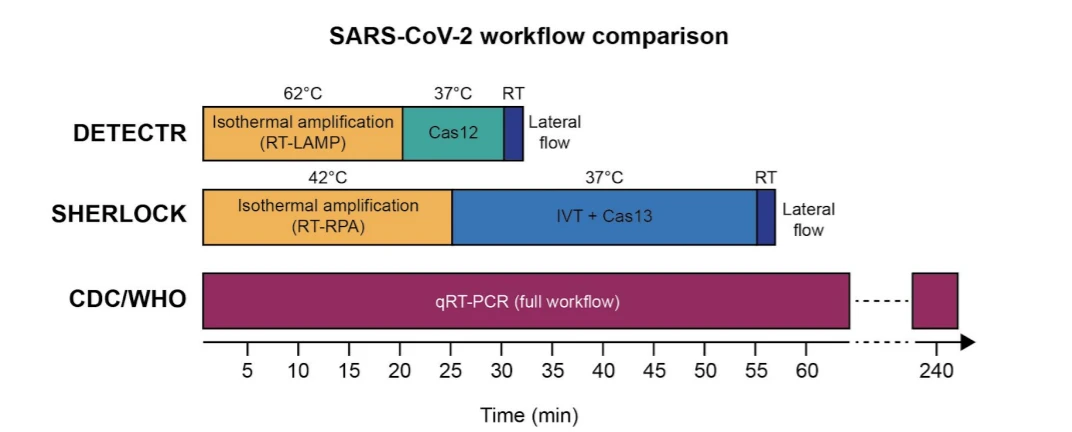
The SHERLOCK detection protocol for COVID-19 virus detection involves three steps:
- Amplification of the synthetic viral RNA using recombinase polymerase amplification (RPA) technology, followed by in vitro transcription of amplified DNA back into RNA.
- RNA-detection using Cas13 nuclease and Synthego-supplied crRNA targeting specific sequences.
- Visual color readout using a commercially-available paper dipstick, which captures the cleaved reporter RNA with labeled ends on specific antibody bands.
The researchers demonstrated that they could use this protocol to detect coronavirus target RNA sequences with a sensitivity of 10-100 copies/µl of input. They note that this technology can be used for testing RNA purified from patient samples in less than an hour without the need for special instrumentation.
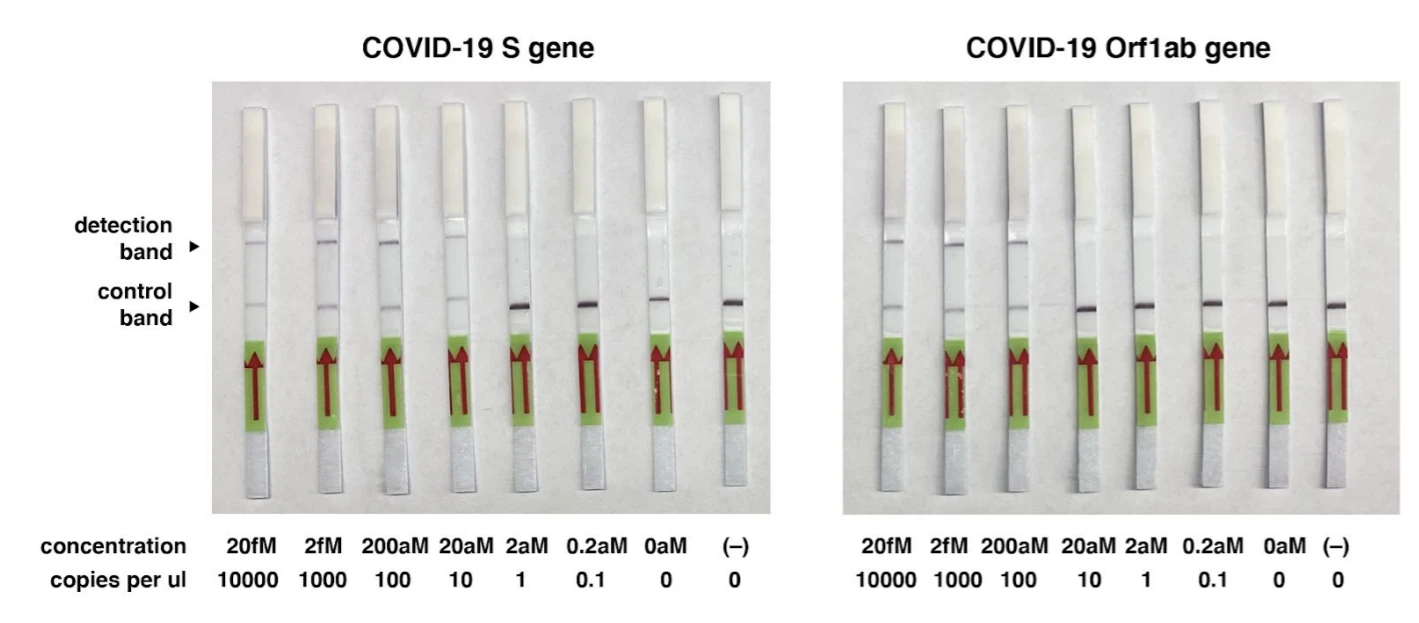
Currently, the protocol has not been validated using actual patient samples but the researchers have noted their availability for guidance in case others wish to perform validation tests.
CRISPR has ignited a revolution. Although it’s a relatively recent discovery in the history of biotechnology, CRISPR has quickly become a standard laboratory tool and cell engineering is transforming research. One of the most widely used applications of CRISPR is knocking out specific genes in cell lines to interrogate gene function.

Mammoth Biosciences Adapts DETECTR Platform for Detection of Coronavirus RNA
Last month, Mammoth Biosciences published a white paper on how the DETECTR platform could be adapted to detect SARS-CoV-2 RNA in about 30 minutes. Such a rapid diagnostic platform would be particularly valuable in high-risk areas such as airports and hospitals.
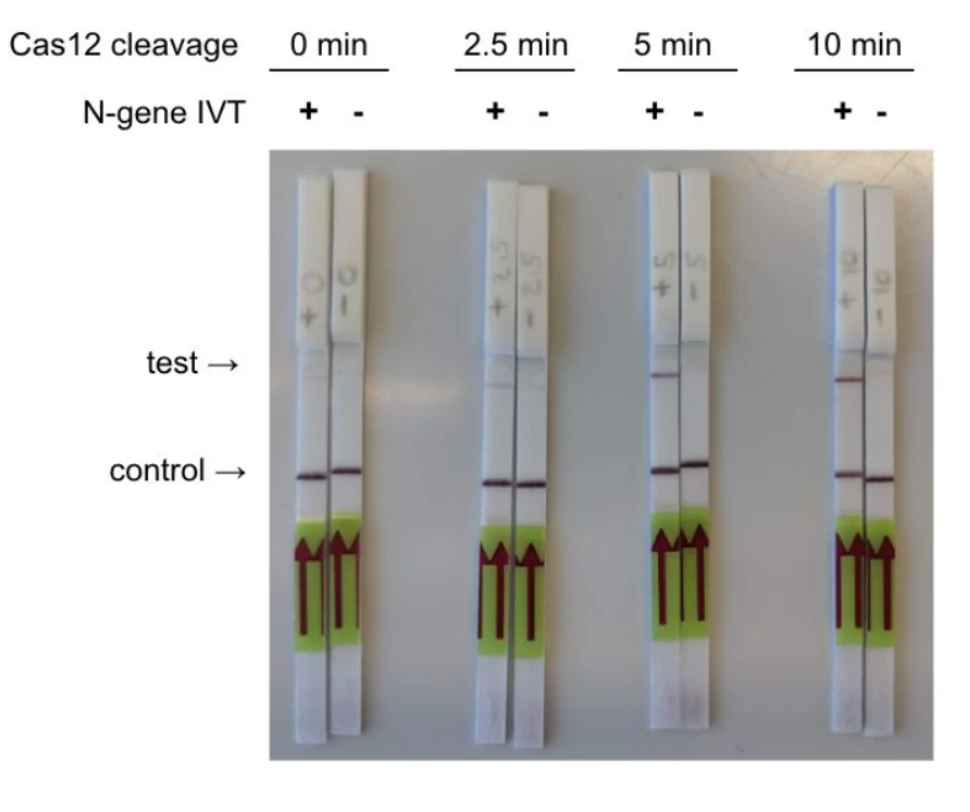
The Mammoth Biosciences team tested CRISPR-based detection of two genes, N-gene and E-gene, from the SARS-CoV-2 genome from contrived samples. The details and results of their test are given in this white paper.
Their protocol involved the following steps:
- Amplification of the RNA extracted from (in this case, contrived) sample using reverse transcription loop-mediated isothermal amplification (RT-LAMP).
- Detection of DNA using Cas12a and Synthego-supplied crRNA targeting specific sequences.
- Visual readout using commercially available dip strips.
Comparing their methods to the SHERLOCK-based protocol, the Mammoth Biosciences team noted that their platform enabled faster detection (30 instead of 60 minutes) as they saved the time spent on the additional IVT steps required for Cas13a-based detection. The sensitivity range of the DETECTR platform is 70-300 copies/µl input.
Synthego’s Commitment to Enable CRISPR Breakthroughs
Synthego’s mission is to enable scientists to make a difference with their discoveries using CRISPR. We are proud to have supported Mammoth Biosciences and Feng Zhang’s team with our high-quality synthetic guide RNAs for use in their diagnostic platforms. We hope to keep partnering with researchers and institutions to support CRISPR diagnostics and therapeutics in the future.
Whether coronavirus becomes a pandemic remains to be seen, but the positive advances in science offer some hope for disease control. In fact, SHERLOCK Biosciences recently announced a partnership with Cepheid to work together on diagnostic platforms for better preparedness in case of future disease outbreaks.